Voronoi: Nodes and Geometry, Integrators
Nodes
The most basic thing is the creation of a list of Points. We advise to use the following:
HighVoronoi.VoronoiNodes — MethodVoronoiNodes(x::Matrix)also available in the forms
VoronoiNodes(x::Vector{<:Vector})
VoronoiNodes(x::Vector{<:SVector})creates a list of points (as static vectors) from a matrix.
Example: 100 Points in $(0,1)^3$
data = rand(3,100)
points = VoronoiNodes(data)An advanced method is given by the following
VoronoiNodes(number_of_nodes::Int;density ,
domain::Boundary=Boundary(), bounding_box::Boundary=Boundary(),
criterium=x->true)When density = x->f(x) this will create a cloud of approximately number_of_nodes points inside the intersection of domain and bounding_box with spatial distribution $f(x)$. Note that both exact number and position of points are random. The variable bounding_box allows to handle also the case when domain is unbounded. The intersection of domain and bounding_box HAS TO BE bounded!
The following two pictures show first a distribution density = x->sin(pi*2*x[1])^2*sin(pi*2*x[2])^2 and the second takes the same density squared.
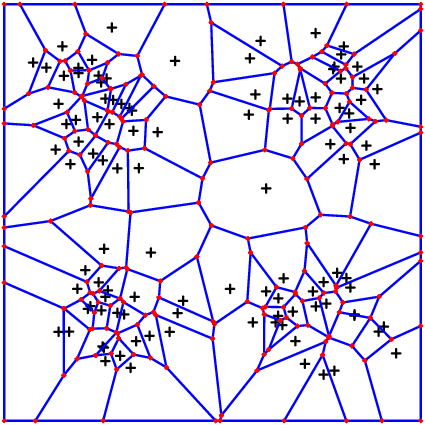
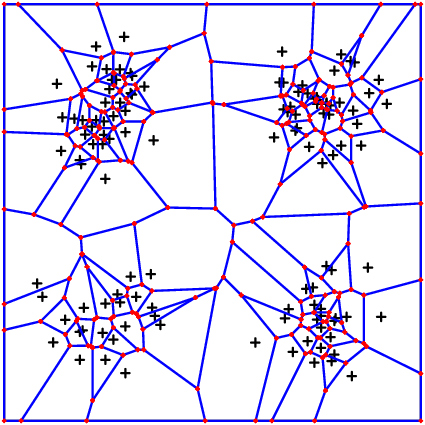
Single Nodes
To instatiate a single node (e.g. if you want to add a specific node to an existing list of nodes) use
# make [1.0, 0.0, 0.5] a valid Voronoi node
VoronoiNode([1.0, 0.0, 0.5])Example
# This is an example to illustrate VoronoiNodes(number_of_nodes::Int;density)
## First some plot routine ############################
using Plots
function plot_2d_surface(nodes, values)
# The following two lines are necessary in order for the plot to look nicely
func = StepFunction(nodes,values)
new_nodes = vcat([VoronoiNode([k/10,j*1.0]) for k in 0:10, j in 0:1], [VoronoiNode([j*1.0,k/10]) for k in 1:9, j in 0:1])
append!(nodes,new_nodes)
append!(values,[func(n) for n in new_nodes])
x = [node[1] for node in nodes]
y = [node[2] for node in nodes]
p = Plots.surface(x, y, values, legend=false)
xlabel!("X")
ylabel!("Y")
zlabel!("Values")
title!("2D Surface Graph")
display(p)
end
########################################################
## Now for the main part ################################
my_distribution = x->(sin(x[1]*π)*sin(x[2]*π))^4
my_nodes = VoronoiNodes(100,density = my_distribution, domain=cuboid(2,periodic=[]))
# you may compare the output to the following:
# my_nodes = VoronoiNodes(100,density = x->1.0, domain=cuboid(2,periodic=[]))
println("This generated $(length(my_nodes)) nodes.")
my_vals = map(x->sin(x[1]*π)^2*sin(x[2]*π),my_nodes)
plot_2d_surface(my_nodes,my_vals)DensityRange
HighVoronoi.DensityRange — TypeDensityRange{S}provides a rectangular grid of points in a S-dimensional space. It is initialized as follows:
DensityRange(mr::AbstractVector{<:Integer},range)Here, range can be of the following types:
AbstractVector{Tuple{<:Real,<:Real}}: It is assumed that each entry ofrangeis a tuple(a_i,b_i)
so the range is defined in the cuboid (a_1,b_1) imes... imes(a_{dim},b_{dim})
mr is assumed to have the same dimension as range and the interval (a_i,b_i) will be devided into mr[i] intervalls
AbstractVector{<:Real}: if e.g.range=[1.0,1.0]this will be transferred torange=[(0.0,1.0),(0.0,1.0)]
and the first instance of the method is called
Float64:rangewill be setrange*ones(Float64,length(mr))and the second instance is calledTuple{<:Real,<:Real}: range will be set to an array of identical tuple entries and the first version is called
Alternatively, one may call the following method:
DensityRange(mr::Int,range,dimension=length(range))it is assumed that range is an array or tuple of correct length and mr is replaced by mr*ones(Int64,dimension). If range is not an array, then dimension has to be provided the correct value.
Geometry
The creation and storage of Voronoi geometry data is handled by the following class.
HighVoronoi.VoronoiGeometry — TypeVoronoiGeometry{T}This is the fundamental struct to store information about the generated Voronoi grid. The geometric data can be accessed using the type VoronoiData. However, there is always the possibility to access the data also via the following fields:
- Integrator.Integral: stores the integrated values in terms of a
Voronoi_Integral - basic_mesh: stores the fundamental data of nodes and verteces. also stored in Integrator.Integral.MESH
- nodes: direct reference to the nodes. Also provided in basic_mesh.nodes
Accessing the data directly, that is without calling VoronoiData, is likely to cause confusion or to provide "wrong" information. The reason is that particularly for periodic boundary conditions, the mesh is enriched by a periodization of the boundary nodes. These nodes are lateron dropped by the VoronoiData-Algorithm.
To create a Voronoi mesh it is most convenient to call either of the following methods
HighVoronoi.VoronoiGeometry — MethodVoronoiGeometry(xs::Points,b::Boundary)This creates a Voronoi mesh from the points xs given e.g. as an array of SVector and a boundary b that might be constructed using the commands in the Boundaries section.
You have the following optional commands:
silence: Suppresses output to the command line whentrue. The latter will speed up the algorithm by a few percent. default isfalse.integrator: can be either one of the following values:VI_GEOMETRY: Only the basic properties of the mesh are provided: the verteces implying a List of neighbors of each nodeVI_MONTECARLO: Volumes, interface areas and integrals are calculated using a montecarlo algorithm. This particular integrator comes up with the following additional paramters:mc_accurate=(int1,int2,int3): Montecarlo integration takes place inint1directions, overint2volumetric samples (vor volume integrals only). It reuses the same set of directionsint3-times to save memory allocation time. Standard setting is:(1000,100,20).
VI_POLYGON: We use the polygon structure of the mesh to calculate the exact values of interface area and volume. The integral over functions is calculated using the values at the center, the verteces and linear interpolation between.VI_HEURISTIC: When this integrator is chosen, you need to provide a fully computed Geometry including volumes and interface areas.VI_HEURISTICwill then use this information to derive the integral values.VI_HEURISTIC_MC: This combines directlyVI_MONTECARLOcalculations of volumes and interfaces and calculates integral values of functions based on those volumes and areas. In particular, it also relies onmc_accurate!
integrand: This is a functionf(x)depending on one spatial variablexreturning aVector{Float64}. The integrated values will be provided for each cell and for each pair of neighbors, i.e. for each interfaceperiodic_grid: This will initiate a special internal routine to fastly create a periodic grid. Look up the section in the documentation.
With density distribution:
VoronoiGeometry(number::Int,b=Boundary();density, kwargs...)this call genertates a distribution of approsximately number nodes and generates a VoronoiGeometry. It takes as parameters all of the above mentioned keywords (though periodic_grid makes no sense) and all keywords valid for a call of VoronoiNodes(number;domain=b,density=density, ....)
In future versions, there will be an implementation of the parameter cubic=true, where the grid will be generated based on a distribution of "cubic" cells. In the current version there will be a warning that this is not yet implemented.
Advanced methods
VoronoiGeometry(file::String)
VoronoiGeometry(VG::VoronoiGeometry)Loads a Voronoi mesh from the file or copies it from the original data VG. If integrator is not provided, it will use the original integrator stored to the file. In the second case, if integrand is not provided explicitly, it will use integrand = VG.integrand as standard. Additionally it has the following options:
_myopen=jldopen: the method to use to open the file. See the section onwrite_jld.offset: See the section onwrite_jld.integrate=false: This will or will not call the integration method after loading/copying the data. Makes sense for usingVI_HEURISTICtogether withvolume=true,area=trueand providing values forintegrandandintegrand. Ifintegrand != nothingbutbulk==falseorinterface==falsethis parameter will internally be settrue.volume=true: Load volume data from filearea=true: Load interface area data from filebulk=false: Load integrated function values on the cell volumes from file. When settrueandintegrand=fis provided the method will compare the dimension offand of the stored data.interface=false: Load integrated function values on the interfaces. When settrueandintegrand=fis provided the method will compare the dimension offand of the stored data.
Integrators (overview)
As discussed above there is a variety of integrators available to the user, plus some internal integrators that we will not discuss in this manual. The important integrators for the user are:
VI_GEOMETRY: Only the basic properties of the mesh are provided: the verteces and an implicit list of neighbors of each node. This is the fastes way to generate aVoronoiGeometryVI_MONTECARLO: Volumes, interface areas and integrals are calculated using a montecarlo algorithm introduced by A. Sikorski inVoronoiGraph.jland discussed in a forthcoming article by Heida, Sikorski, Weber. This particular integrator comes up with the following additional paramters:mc_accurate=(int1,int2,int3): Montecarlo integration takes place inint1directions, overint2volumetric samples (vor volume integrals only). It reuses the same set of directionsint3-times to save memory allocation time. Standard setting is:(1000,100,20).
VI_POLYGON: We use the polygon structure of the mesh to calculate the exact values of interface area and volume. The integral over functions is calculated using the values at the center, the verteces and linear interpolation between. Also this method is to be discussed in the anounced article by Heida, Sikorski, Weber.VI_HEURISTIC: When this integrator is chosen, you need to provide a fully computed Geometry including volumes and interface areas.VI_HEURISTICwill then use this information to derive the integral values.VI_HEURISTIC_MC: This combines directlyVI_MONTECARLOcalculations of volumes and interfaces and calculates integral values of functions based on those volumes and areas. In particular, it also relies onmc_accurate!
It is important to have in mind that the polygon-integrator will be faster in low dimensions, whereas the Montecarlo integrator will outperform from 5 dimensions and higher. However, when volumes and integrals are to be calculated in high dimensions, the VI_HEURISTIC_MC is highly recommended, as it works with much less function evaluations than the VI_MONTECARLO.
Storage
HighVoronoi.write_jld — MethodThe data can be stored using the write_jld method:
write_jld(Geo::VoronoiGeometry,filename,offset="";_myopen=jldopen)
write_jld(Geo::VoronoiGeometry,file,offset="")stores the complete information of a VoronoiGeometry object to a file. This information can later be retrieved using the VoronoiGeometry(file::String, args...) function.
Geo: The Voronoi geometry object to be storedfilename: name of file to store infile: A file given in a format supportingwrite(file,"tagname",content)andread(file,"tagname",content)offset: If several Geometry objects are to be stored in the same file, this will be the possibility to identify each one by a unique name. In particular, this is the key to store several objects in one single file._myopen: a method that allows the syntax_myopen(filename,"w") do myfile ....... end. By default the method uses theJLD2library as this (at the point of publishing this package) has the least problems with converting internal data structure to an output format.
If you want to use the default method, then the filename should end on .jld. Otherwise there might be confusion by the abstract built in julia loading algorithm.
HighVoronoi.load_Voronoi_info — MethodIf you want to make sure that the data you load will be rich enough, compact information can be retrieved as follows:
load_Voronoi_info(filname,offset="")This will print out compact information of the data stored in file filename and the offset offset. Yields dimension, number of nodes, number of internal nodes and the dimension of the stored integrated data. Note that the latter information is of particular importance since here is the highest risk for the user to mess up stored data with the algorithm.
Extraction of VoronoiData data for further processing
HighVoronoi.VoronoiData — TypeUsing the call
data=VoronoiData(VG)some data of the Voronoi geometry VG is extracted. Once applied, the data set contains at least the following informations:
nodes::Vector{T}: The original nodesneighbors::Vector{Vector{Int64}}: For each nodenodes[i]the fieldneighbors[i]contains a sorted list of indeces of all neighboring cells. Multiple appearence of the same node is possible on a periodic grid.
Fields in VoronoiData
Conditionally on what the VoronoiGeometry VG was told to calculate, the set data contains the following additional information:
volume::Vector{Float64}: the volume for each nodearea::Vector{Vector{Float64}}: stores for each neighborneighbors[i][k]of nodeiinarea[i][k]the area of the interface.bulk_integral::Vector{Vector{Float64}}: the integral over the bulk functioninterface_integral::Vector{Vector{Vector{Float64}}}: same as forareabut with the integral values of the interface function. In paricularinterface_integral[i][k]is of typeVector{Float64}
If the four above data fieds where not requested to be calculated, the vectors have length 0 and any attempt to access their values will eventually result in an error message.
Named Arguments
The call of VoronoiData(VG) provides the following options:
view_only=false: Iftruethis implies that fornodes,volume,area,bulk_integralandinterface_integralonly views on the internally stored data will be provided. When set tofalse, deep copies of the of the internal data will be provided.neighborswill always be explicitly defined within this dataset only (i.e. noview)reduce_to_periodic=true: This erases all data generated from the periodization. It is highly advised to set this option totrueas the user will then only see the periodic mesh as one would expect it.getorientations=false: This is set automatically totrueoncereduce_to_periodic==true. Once settruethe result contains the fieldorientations::Vector{Vector{T}},Tbeing the type of d-dimensional vectors originally provided by the nodes of the grid.getvertices=false: Set totruethe fieldverteces::Vector{Dict{Vector{Int64},T}}will for each nodeicontain a dictionary[nodes]=>coordinate. furthermore, the fieldboundary_verteces::Dict{Vector{Int64},boundary_vertex{T}}will contain a list of edges that go to infinitygetboundarynodes=false: Set totruethe fieldboundary_nodes::Dict{Int64,Dict{Int64,T}}will contain a dictionarynode=>Dict(boundary=>point), where boundary is the indexboundary = length(nodes) + number_of_boundary_plane. Whenonboundary==falsethenpointwill be mirrored version ofnodes[node]at the boundary planenumber_of_boundary_plane. Otherwise,pointis the center ofnodes[node]and its mirrored versiononboundary=false: Explained in the last topic. Furthermore, the value is stored inboundary_nodes_on_boundary::Boolsorted=true: During the reduction of the internal pseude periodic mesh to the fully periodic output, the neighbors (jointly with their respective properties) get sorted by their numbers