xxxxxxxxxxbegin using Pkg; Pkg.activate(mktempdir()) Pkg.add("PlutoUI") Pkg.add("PyPlot") Pkg.add("ExtendableSparse") Pkg.add("BenchmarkTools") using PlutoUI,PyPlot,BenchmarkToolsend;pyplot (generic function with 1 method)xxxxxxxxxx# A function to handle sizing and return of a pyplot figurefunction pyplot(f;width=3,height=3) clf() f() fig=gcf() fig.set_size_inches(width,height) fig endSparse matrices
In the previous lectures we found examples of matrices from partial differential equations which have only 3 of 5 nonzero diagonals. For 3D computations this would be 7 diagonals. One can make use of this diagonal structure, e.g. when coding the progonka method.
Matrices from unstructured meshes for finite element or finite volume methods have a more irregular pattern, but as a rule only a few entries per row compared to the number of unknowns. In this case storing the diagonals becomes unfeasible.
Definition: We call a matrix sparse if regardless of the number of unknowns
If we find a scheme which allows to store only the non-zero matrix entries, we would need not more than
The same would be true for the matrix-vector multiplication if we program it in such a way that we use every nonzero element just once: matrix-vector multiplication would use
What is a good storage format for sparse matrices?
Is there a way to implement Gaussian elimination for general sparse matrices which allows for linear system solution with
Is there a way to implement Gaussian elimination \emph{with pivoting} for general sparse matrices which allows for linear system solution with
Is there any algorithm for sparse linear system solution with
Triplet storage format
Store all nonzero elements along with their row and column indices
One real, two integer arrays, length = nnz= number of nonzero elements
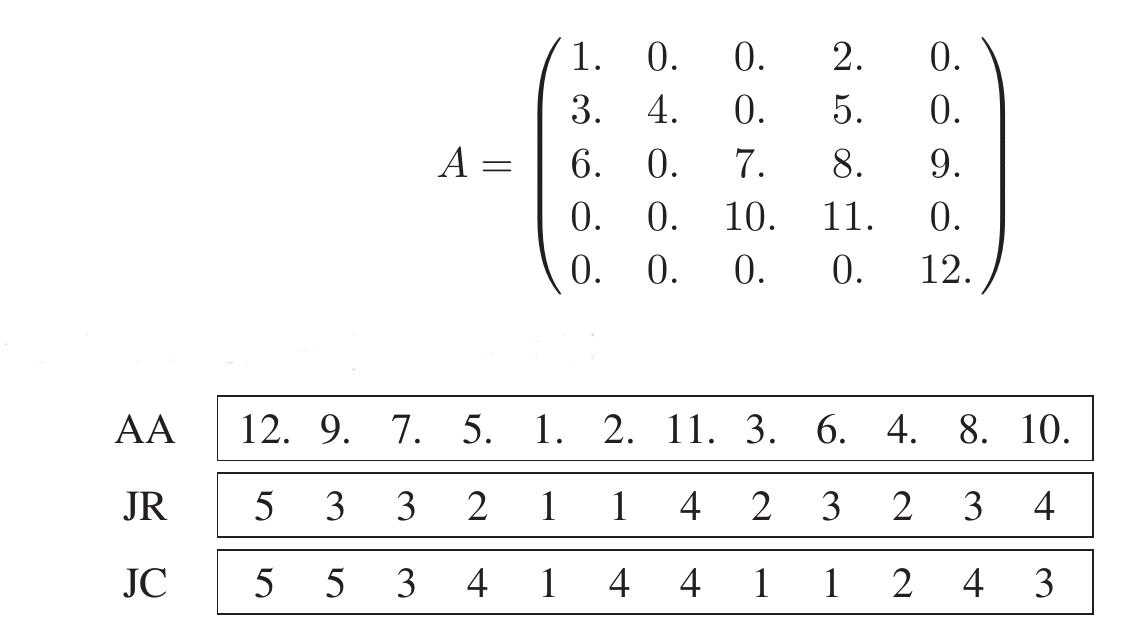
(Y.Saad, Iterative Methods, p.92)
Also known as Coordinate (COO) format
This format often is used as an intermediate format for matrix construction
Compressed Sparse Row (CSR) format
(aka Compressed Sparse Row (CSR) or IA-JA etc.)
float array
AA, length nnz, containing all nonzero elements row by rowinteger array
JA, length nnz, containing the column indices of the elements ofAAinteger array
IA, length N+1, containing the start indizes of each row in the arraysIAandJAandIA[N+1]=nnz+1
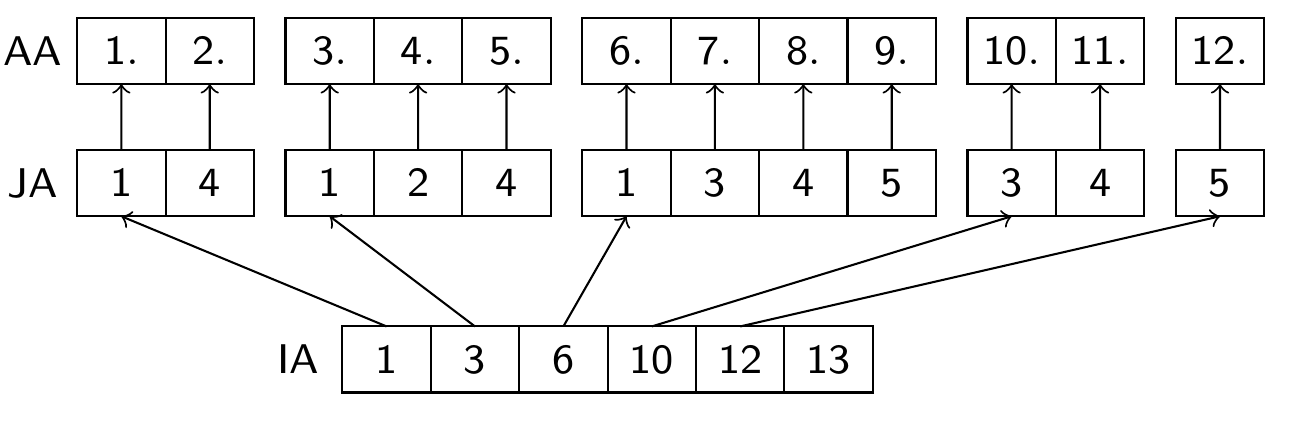
Used in many sparse matrix solver packages
Compressed Sparse Column (CSC) format
Uses similar principle but stores the matrix column-wise.
Used in Julia
Sparse matrices in Julia
xxxxxxxxxxusing SparseArrays,LinearAlgebraCreate sparse matrix from a full matrix
5×5 Array{Float64,2}:
1.0 0.0 0.0 2.0 0.0
3.0 4.0 0.0 5.0 0.0
6.0 0.0 7.0 8.0 9.0
0.0 0.0 10.0 11.0 0.0
0.0 0.0 0.0 0.0 12.0A=Float64[1 0 0 2 0; 3 4 0 5 0; 6 0 7 8 9; 0 0 10 11 0; 0 0 0 0 12]5×5 SparseMatrixCSC{Float64,Int64} with 12 stored entries:
[1, 1] = 1.0
[2, 1] = 3.0
[3, 1] = 6.0
[2, 2] = 4.0
[3, 3] = 7.0
[4, 3] = 10.0
[1, 4] = 2.0
[2, 4] = 5.0
[3, 4] = 8.0
[4, 4] = 11.0
[3, 5] = 9.0
[5, 5] = 12.0As=sparse(A)1
4
5
7
11
13
xxxxxxxxxxAs.colptr1
2
3
2
3
4
1
2
3
4
3
5
xxxxxxxxxxAs.rowval1.0
3.0
6.0
4.0
7.0
10.0
2.0
5.0
8.0
11.0
9.0
12.0
xxxxxxxxxxAs.nzvalpyplot(width=2,height=2) do spy(As,marker=".")endCreate a random sparse matrix
100xxxxxxxxxxN=1000.1xxxxxxxxxxp=0.1Random sparse matrix with probability p=0.1 that
100×100 SparseMatrixCSC{Float64,Int64} with 1032 stored entries:
[7 , 1] = 0.94222
[11, 1] = 0.0328404
[14, 1] = 0.288891
[27, 1] = 0.777506
[29, 1] = 0.499039
[38, 1] = 0.493717
⋮
[3 , 100] = 0.209442
[7 , 100] = 0.52545
[25, 100] = 0.570221
[38, 100] = 0.360959
[61, 100] = 0.174752
[77, 100] = 0.862321
[86, 100] = 0.562663xxxxxxxxxxA2=sprand(N,N,p)xxxxxxxxxxpyplot(width=3,height=3) do spy(A2,marker=".",markersize=0.5)endCreate a sparse matrix from given data
There are several possibilities to create a sparse matrix for given data
As an example, we create a tridiagonal matrix.
10000N1=100000.178295
0.737103
0.370098
0.837115
0.313983
0.349467
0.546892
0.72494
0.929888
0.700349
0.254991
0.366964
0.332019
0.935532
0.503325
0.735556
0.17577
0.42256
0.841844
0.340334
0.12482
0.0863661
0.175945
0.917981
0.895322
0.193474
0.945499
0.909567
0.901804
0.267438
xxxxxxxxxxa=rand(N1-1)0.333554
0.378649
0.622097
0.0677654
0.230456
0.348583
0.495553
0.74695
0.12383
0.328722
0.769973
0.218627
0.995542
0.338912
0.929446
0.682969
0.0619503
0.0615952
0.480887
0.478574
0.653152
0.879161
0.290712
0.392596
0.915582
0.183284
0.815526
0.65137
0.894436
0.647475
xxxxxxxxxxb=rand(N1)0.604986
0.210584
0.889549
0.725572
0.147618
0.784768
0.793034
0.26308
0.302458
0.391863
0.031989
0.820751
0.0581305
0.971845
0.608143
0.239522
0.741068
0.658797
0.237352
0.536843
0.92996
0.541662
0.573119
0.193352
0.799961
0.0674989
0.450361
0.663642
0.944804
0.0753511
xxxxxxxxxxc=rand(N1-1)Special case: use the Julia tridiagonal matrix constructor
sptri_special (generic function with 1 method)sptri_special(a,b,c)=sparse(Tridiagonal(a,b,c))Create an empty Julia sparse matrix and fill it incrementally
10×10 SparseMatrixCSC{Float64,Int64} with 0 stored entriesxxxxxxxxxxB=spzeros(10,10)3xxxxxxxxxxB[1,2]=310×10 SparseMatrixCSC{Float64,Int64} with 1 stored entry:
[1, 2] = 3.0xxxxxxxxxxBsptri_incremental (generic function with 1 method)xxxxxxxxxxfunction sptri_incremental(a,b,c) N=length(b) A=spzeros(N,N) A[1,1]=b[1] A[1,2]=c[1] for i=2:N-1 A[i,i-1]=a[i-1] A[i,i]=b[i] A[i,i+1]=c[i] end A[N,N-1]=a[N-1] A[N,N]=b[N] AendUse the coordinate format as intermediate storage, and construct sparse matrix from there. This is the recommended way.
sptri_coo (generic function with 1 method)function sptri_coo(a,b,c) N=length(b) II=[1,1] JJ=[1,2] AA=[b[1],c[1]] for i=2:N-1 push!(II,i) push!(JJ,i-1) push!(AA,a[i-1]) push!(II,i) push!(JJ,i) push!(AA,b[i]) push!(II,i) push!(JJ,i+1) push!(AA,c[i]) end push!(II,N) push!(JJ,N-1) push!(AA,a[N-1]) push!(II,N) push!(JJ,N) push!(AA,b[N]) sparse(II,JJ,AA)endUse the ExtendableSparse.jl package which implicitely uses the so-called linked list format for intermediate storage of new entries. Note the flush!() method which needs to be called in order to transfer them to the Julia sparse matrix structure.
xxxxxxxxxxusing ExtendableSparsesptri_ext (generic function with 1 method)xxxxxxxxxxfunction sptri_ext(a,b,c) N=length(b) A=ExtendableSparseMatrix(N,N) A[1,1]=b[1] A[1,2]=c[1] for i=2:N-1 A[i,i-1]=a[i-1] A[i,i]=b[i] A[i,i+1]=c[i] end A[N,N-1]=a[N-1] A[N,N]=b[N] flush!(A)endBenchmarkTools.Trial:
memory estimate: 547.27 KiB
allocs estimate: 8
--------------
minimum time: 38.350 μs (0.00% GC)
median time: 42.076 μs (0.00% GC)
mean time: 58.949 μs (19.93% GC)
maximum time: 1.513 ms (94.22% GC)
--------------
samples: 10000
evals/sample: 1xxxxxxxxxx sptri_special(a,b,c)BenchmarkTools.Trial:
memory estimate: 1.08 MiB
allocs estimate: 33
--------------
minimum time: 18.266 ms (0.00% GC)
median time: 18.774 ms (0.00% GC)
mean time: 18.830 ms (0.11% GC)
maximum time: 20.520 ms (0.00% GC)
--------------
samples: 266
evals/sample: 1xxxxxxxxxx sptri_incremental(a,b,c)BenchmarkTools.Trial:
memory estimate: 2.65 MiB
allocs estimate: 66
--------------
minimum time: 621.986 μs (0.00% GC)
median time: 647.085 μs (0.00% GC)
mean time: 727.777 μs (7.74% GC)
maximum time: 2.324 ms (60.01% GC)
--------------
samples: 6861
evals/sample: 1xxxxxxxxxx sptri_coo(a,b,c)BenchmarkTools.Trial:
memory estimate: 1.53 MiB
allocs estimate: 25
--------------
minimum time: 681.731 μs (0.00% GC)
median time: 740.557 μs (0.00% GC)
mean time: 784.821 μs (4.09% GC)
maximum time: 2.394 ms (63.66% GC)
--------------
samples: 6368
evals/sample: 1xxxxxxxxxx sptri_ext(a,b,c)Benchmark summary:
The incremental creation of a SparseMartrixCSC from an initial state with non nonzero entries is slow because of the data shifts and reallocations necessary during the construction
The COO intermediate format is sufficiently fast, but inconvenient
The ExtendableSparse package provides has similar peformance and is easy to use.
Sparse direct solvers
Sparse direct solvers implement LU factorization with different pivoting strategies. Some examples:
UMFPACK: e.g. used in Julia
Pardiso (omp + MPI parallel)
SuperLU (omp parallel)
MUMPS (MPI parallel)
Pastix
Quite efficient for 1D/2D problems - we will discuss this more deeply
Essentially they implement the LU factorization algorithm
They suffer from fill-in, especially for 3D problems:
Let
increased memory usage to store L,U
high operation count
pyplot(width=3,height=3) do spy(A2,marker=".",markersize=0.5)endpyplot(width=3,height=3) do spy(lu(A2).L,marker=".",markersize=0.5)endpyplot(width=3,height=3) do spy(lu(A2).U,marker=".",markersize=0.5)end1032
3783
nnz(A2), nnz(lu(A2))Solution steps with sparse direct solvers
Pre-ordering
Decrease amount of non-zero elements generated by fill-in by re-ordering of the matrix
Several, graph theory based heuristic algorithms exist
Symbolic factorization
If pivoting is ignored, the indices of the non-zero elements are calculated and stored
Most expensive step wrt. computation time
Numerical factorization
Calculation of the numerical values of the nonzero entries
Moderately expensive, once the symbolic factors are available
Upper/lower triangular system solution
Fairly quick in comparison to the other steps
Separation of steps 2 and 3 allows to save computational costs for problems where the sparsity structure remains unchanged, e.g. time dependent problems on fixed computational grids
With pivoting, steps 2 and 3 have to be performed together, and pivoting can increase fill-in
Instead of pivoting, iterative refinement may be used in order to maintain accuracy of the solution
Influence of reordering
Sparsity patterns for original matrix with three different orderings of unknowns
number of nonzero elements (of course) independent of ordering:
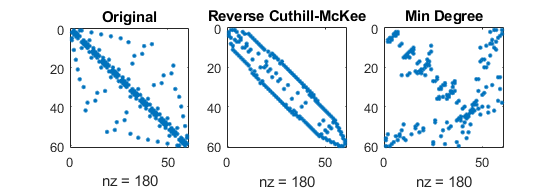
(mathworks.com)
Sparsity patterns for corresponding LU factorizations
number of nonzero elements depend original ordering!
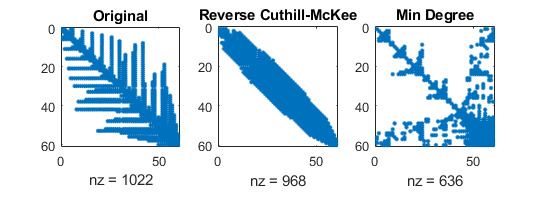
(mathworks.com)
Sparse direct solvers: Complexity estimate
Complexity estimates depend on storage scheme, reordering etc.
Sparse matrix - vector multiplication has complexity
Some estimates can be given from graph theory for discretizations of heat equation with
sparse LU factorization:
triangular solve: work dominated by storage complexity
(Source: J. Poulson, PhD thesis)
Practical use
\operator
Asparse_incr=sptri_incremental(a,b,c);7.3839
-2.41811
2.84497
1.13828
-0.17924
0.599098
1.07987
0.322186
0.641531
2.27137
-0.875819
2.61354
0.794315
-12.8994
5.25603
13.4551
-45.2354
-8.2241
14.3558
-10.2311
2.49771
-2.57255
5.31473
-0.478023
-4.61416
83.744
-29.8787
-81.0875
85.7264
-33.8646
x
Asparse_incr\ones(N1)10000×10000 ExtendableSparseMatrix{Float64,Int64}:
0.333554 0.604986 0.0 0.0 … 0.0 0.0 0.0 0.0
0.178295 0.378649 0.210584 0.0 0.0 0.0 0.0 0.0
0.0 0.737103 0.622097 0.889549 0.0 0.0 0.0 0.0
0.0 0.0 0.370098 0.0677654 0.0 0.0 0.0 0.0
0.0 0.0 0.0 0.837115 0.0 0.0 0.0 0.0
0.0 0.0 0.0 0.0 … 0.0 0.0 0.0 0.0
0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0
⋮ ⋱
0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0
0.0 0.0 0.0 0.0 … 0.450361 0.0 0.0 0.0
0.0 0.0 0.0 0.0 0.815526 0.663642 0.0 0.0
0.0 0.0 0.0 0.0 0.909567 0.65137 0.944804 0.0
0.0 0.0 0.0 0.0 0.0 0.901804 0.894436 0.0753511
0.0 0.0 0.0 0.0 0.0 0.0 0.267438 0.647475xxxxxxxxxxAsparse_ext=sptri_ext(a,b,c)7.3839
-2.41811
2.84497
1.13828
-0.17924
0.599098
1.07987
0.322186
0.641531
2.27137
-0.875819
2.61354
0.794315
-12.8994
5.25603
13.4551
-45.2354
-8.2241
14.3558
-10.2311
2.49771
-2.57255
5.31473
-0.478023
-4.61416
83.744
-29.8787
-81.0875
85.7264
-33.8646
Asparse_ext\ones(N1)